Apologies for the long lapse in response here, it's been a busy few months! Giulio Lampronti and I spend a bit of time looking at this back in December, and managed to get it working well in the input file itself. However, I'm finding a little difficulty in converting to the form of a macro. The code that works (at least for monoclinic, it shouldn't be difficult to change for the rest) is as follows: (note, the forums may format this slightly dodgy, so have also attached screenshots)
prm s400 1e+161.05335787 min 0.000000000000001 max 1e+20
prm s040 4.02030236e+10_1.05335787_LIMIT_MIN_1e-15 min 0.000000000000001 max 1e+20
prm s004 8.03640459e+102.78826643e+20_LIMIT_MIN_1e-15 min 0.000000000000001 max 1e+20
prm s220 8.28938399e+12_1.05335787 max 1e+20
prm s202 -1.23829058e+121.05335787 max 1e+20
prm s022 1.60728092e+11_1.05335787 max 1e+20
prm s301 3.48795127e+091.05335787 max 1e+20
prm s121 2.40541613e+11_1.05335787 max 1e+20
prm s103 -9.14581569e+111.05335787 max 1e+20
prm eta_TOF 0.999999871`0.210782633_LIMIT_MIN_1e-10 min 0.0000000001 max 0.9999999999
prm mhkl = H4 s400 + K4 s040 + L4 s004
+ H2 K2 s220 + H2 L2 s202 + K2 L2 s022
+ H3 L1 s301 + H1 L3 s103 + H1 K2 L1 s121;
'lor_fwhm = eta * (D_spacing^2 * Sqrt(Max(mhkl,0)) Tan(Th) 0.0018/3.1415927;
'gauss_fwhm = (1 - eta) * (D_spacing^2 * Sqrt(Max(mhkl,0)) Tan(Th) 0.0018/3.1415927; `

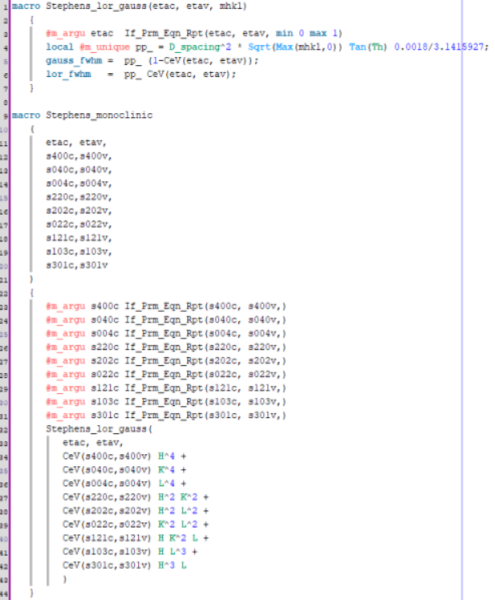
To adapt this into a macro, I attempted to use the XRD macro from our topas.inc as inspiration:
`macro Stephens_lor_gauss(etac, etav, mhkl)
{
#m_argu etac If_Prm_Eqn_Rpt(etac, etav, min 0 max 1)
local #m_unique pp_ = D_spacing2 * Sqrt(Max(mhkl,0)) Tan(Th) 0.0018/3.1415927;
gauss_fwhm = pp_ (1-CeV(etac, etav));
lor_fwhm = pp_ CeV(etac, etav);
}
macro Stephens_monoclinic
(
etac, etav,
s400c,s400v,
s040c,s040v,
s004c,s004v,
s220c,s220v,
s202c,s202v,
s022c,s022v,
s121c,s121v,
s103c,s103v,
s301c,s301v
)
{
#m_argu s400c If_Prm_Eqn_Rpt(s400c, s400v,)
#m_argu s040c If_Prm_Eqn_Rpt(s040c, s040v,)
#m_argu s004c If_Prm_Eqn_Rpt(s004c, s004v,)
#m_argu s220c If_Prm_Eqn_Rpt(s220c, s220v,)
#m_argu s202c If_Prm_Eqn_Rpt(s202c, s202v,)
#m_argu s022c If_Prm_Eqn_Rpt(s022c, s022v,)
#m_argu s121c If_Prm_Eqn_Rpt(s121c, s121v,)
#m_argu s103c If_Prm_Eqn_Rpt(s103c, s103v,)
#m_argu s301c If_Prm_Eqn_Rpt(s301c, s301v,)
Stephens_lor_gauss(
etac, etav,
CeV(s400c,s400v) H4 +
CeV(s040c,s040v) K4 +
CeV(s004c,s004v) L4 +
CeV(s220c,s220v) H2 K2 +
CeV(s202c,s202v) H2 L2 +
CeV(s022c,s022v) K2 L2 +
CeV(s121c,s121v) H K2 L +
CeV(s103c,s103v) H L3 +
CeV(s301c,s301v) H3 L
)
}
`
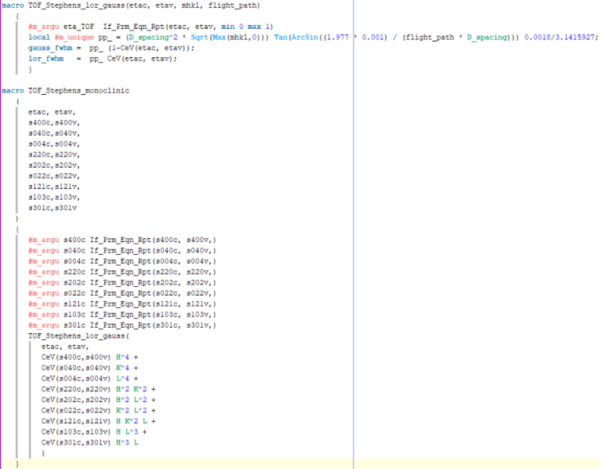
Below is as far as I've gotten with the TOF macro. I think the main issue is that I need to incorporate the flightpath variable which is now present in the stephens_lor_gauss function, which I'm not quite sure how to do:
`macro TOF_Stephens_lor_gauss(etac, etav, mhkl, flight_path)
{
#m_argu eta_TOF If_Prm_Eqn_Rpt(etac, etav, min 0 max 1)
local #m_unique pp_ = (D_spacing2 * Sqrt(Max(mhkl,0))) Tan(ArcSin((1.977 * 0.001) / (flight_path * D_spacing))) 0.0018/3.1415927;
gauss_fwhm = pp_ (1-CeV(etac, etav));
lor_fwhm = pp_ CeV(etac, etav);
}
macro TOF_Stephens_monoclinic
(
etac, etav,
s400c,s400v,
s040c,s040v,
s004c,s004v,
s220c,s220v,
s202c,s202v,
s022c,s022v,
s121c,s121v,
s103c,s103v,
s301c,s301v
)
{
#m_argu s400c If_Prm_Eqn_Rpt(s400c, s400v,)
#m_argu s040c If_Prm_Eqn_Rpt(s040c, s040v,)
#m_argu s004c If_Prm_Eqn_Rpt(s004c, s004v,)
#m_argu s220c If_Prm_Eqn_Rpt(s220c, s220v,)
#m_argu s202c If_Prm_Eqn_Rpt(s202c, s202v,)
#m_argu s022c If_Prm_Eqn_Rpt(s022c, s022v,)
#m_argu s121c If_Prm_Eqn_Rpt(s121c, s121v,)
#m_argu s103c If_Prm_Eqn_Rpt(s103c, s103v,)
#m_argu s301c If_Prm_Eqn_Rpt(s301c, s301v,)
TOF_Stephens_lor_gauss(
etac, etav,
CeV(s400c,s400v) H4 +
CeV(s040c,s040v) K4 +
CeV(s004c,s004v) L4 +
CeV(s220c,s220v) H2 K2 +
CeV(s202c,s202v) H2 L2 +
CeV(s022c,s022v) K2 L2 +
CeV(s121c,s121v) H K2 L +
CeV(s103c,s103v) H L3 +
CeV(s301c,s301v) H3 L
)
} `
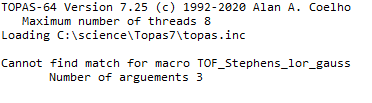
In the input file itself, flight_path is defined locally for each bank, is it possible to tell the macro just to take the local flightpath macro? If so, how do I direct it to do this?

Thanks again all for your continued help here! Hopefully we'll have a set of TOF_Stephens strain macros to share with the TOPAS community soon!