dear Prof Evans - thanks for your reply.
I believe space group to be correct (Fdd2).
MVW output looks OK: (MVW( 4675.947, 4524.199, 100.000))
View structure looks OK, but when I apply labels (as you sugggest) I think there is a problem with one component of the cocrystal, it seems like there are 2 labels for each atomic position:
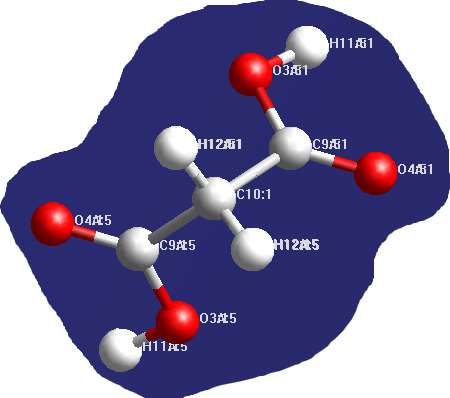
num_positions shows 16 for all apart from C10 position (num_positions = 8) and this atom lies on a 2-fold axis
If half my malonic acid molecule is symmetry generated, what is the best way to deal with this in Topas for the refinement? I have changed the occupancy of each position to 0.5 - is this the most appropriate way to sort it out?
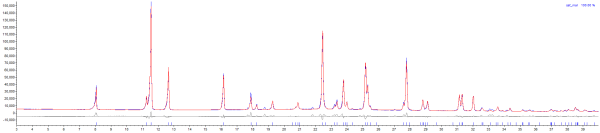
Fit is now far better: